Publications
Quantifying organellar ultrastructure in cryo-electron tomography using a surface morphometrics pipeline.
Barad BA†, Medina M†, Fuentes D, Wiseman, RL, Grotjahn DA. Journal of Cell Biology 2023 †Equal contribution
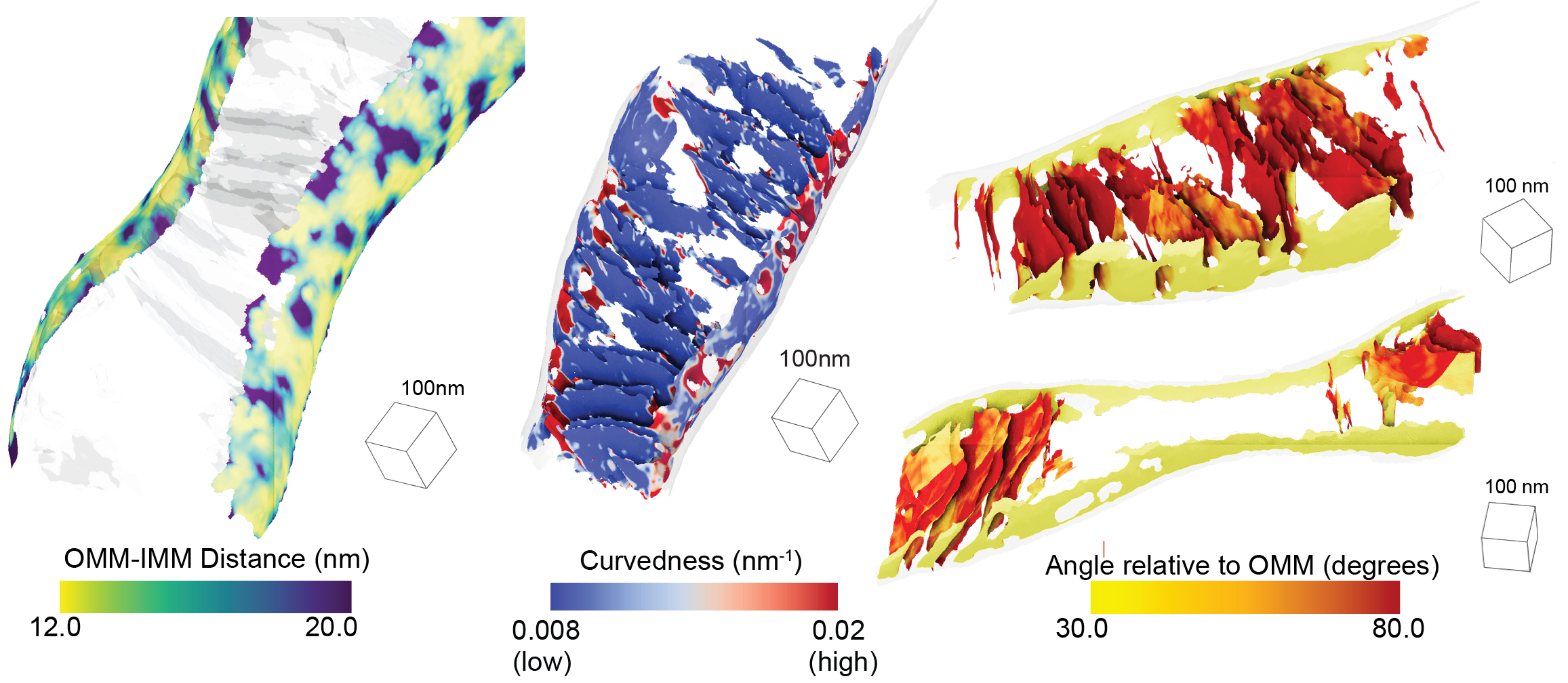
-
Access the paper
- PMID: 36786771
- Biorxiv Preprint: 477440
- Full Text Additional Linkss:
- Surface Morphometrics on Github
- EMPIAR Deposition (Tilt Series, Tomograms, Segmentations, and Surfaces)
- Zenodo Deposition (Quantifications and Statistics)
- Review Commons Reviews
- Wiseman Lab @ Scripps Research
- Preprint Tweetorial
- Journal Tweetorial
Mapping Protein Dynamics at High-Resolution with Temperature-Jump X-ray Crystallography
Wolff AM, Nango E, Young ID, Brewster AS, Kubo M, Nomura T, Sugahara M, Owada S, Barad BA, Ito K, Bhowmick A, Carbajo S, Hino T, Holton JM, Im D, O’Riordan LJ, Tanaka T, Tanaka R, Sierra RG, Yumoto F, Tono K, Iwata S, Sauter NK, Fraser JS, Thompson MC. (2022) bioRxiv.
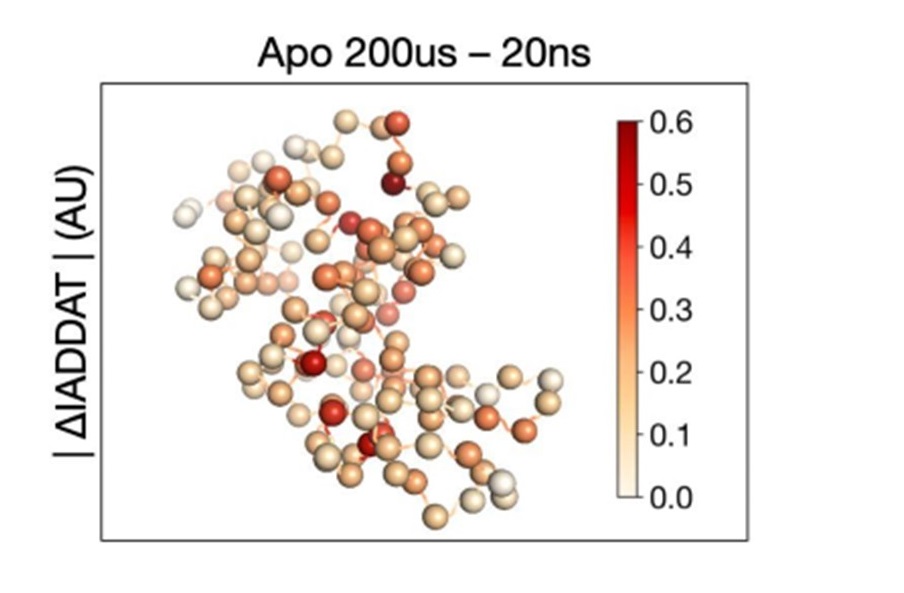
-
Access the paper
- Biorxiv Preprint: 495662
- Full Text
- Online Dataset: doi:10.11577/1873469
- Deposited Structures: 8CVU, 8CVV, 8CVW, 8CW0, 8CW1, 8CW3, 8CW5, 8CW6, 8CW7, 8CW8, 8CWB, 8CWC, 8CWD, 8CWE, 8CWF, 8CWG, 8CWH Additional Linkss:
- Temperature Analysis on Github
- Nango lab @ Tohoku University
- Fraser lab @ UC San Francisco
- Thompson Lab @ UC Merced
- SACLA XFEL
- BioSciences @ LBNL
- Preprint Tweetstorm
Nanoscale details of mitochondrial fission revealed by cryo-electron tomography.
Mageswaran, SK†, Grotjahn DA†§, Zeng X, Barad BA, Medina M, Hoang MH, Dobro MJ, Chang YW, Xu M, Yang WY, Jensen, GJ§. bioRxiv 2021 †Equal contribution §Corresponding authors
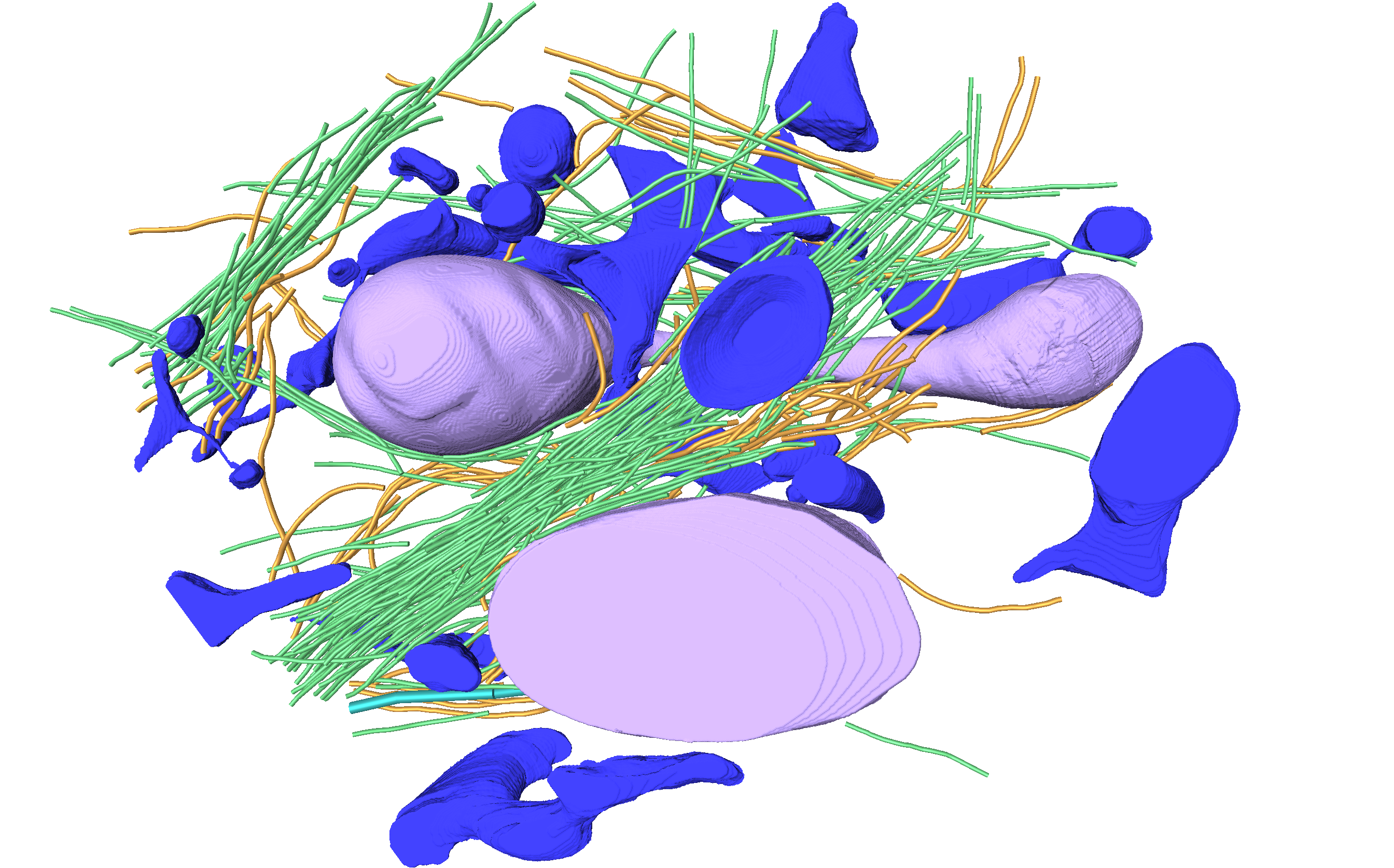
-
Access the paper
- Biorxiv Preprint: 472487
- Full Text Additional Linkss:
- Filament Architecture on Github
- Jensen Lab
CellPAINT: Turnkey illustration of molecular cell biology
Gardner A, Ludovic A, Fuentes D, Maritan M, Barad BA, Medina, M, Olson AJ, Grotjahn DA, Goodsell DS. Front. Bioinform. 2021.
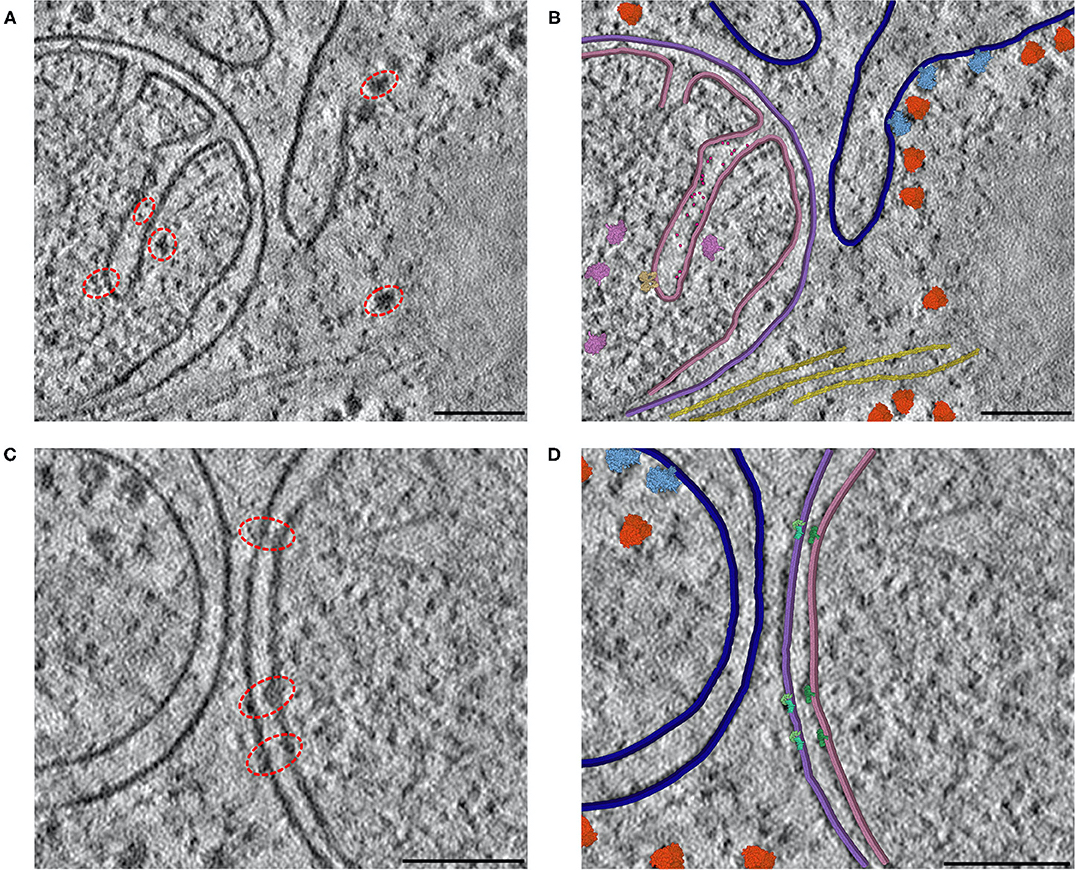
-
Access the paper
- PMID: 34790910
- PMCID: PMC8594902
- Full Text
- Zenodo Record:
(Tomogram used for CellPAINT hypothesis generation)
Additional Links:
- Molecular Graphics Laboratory
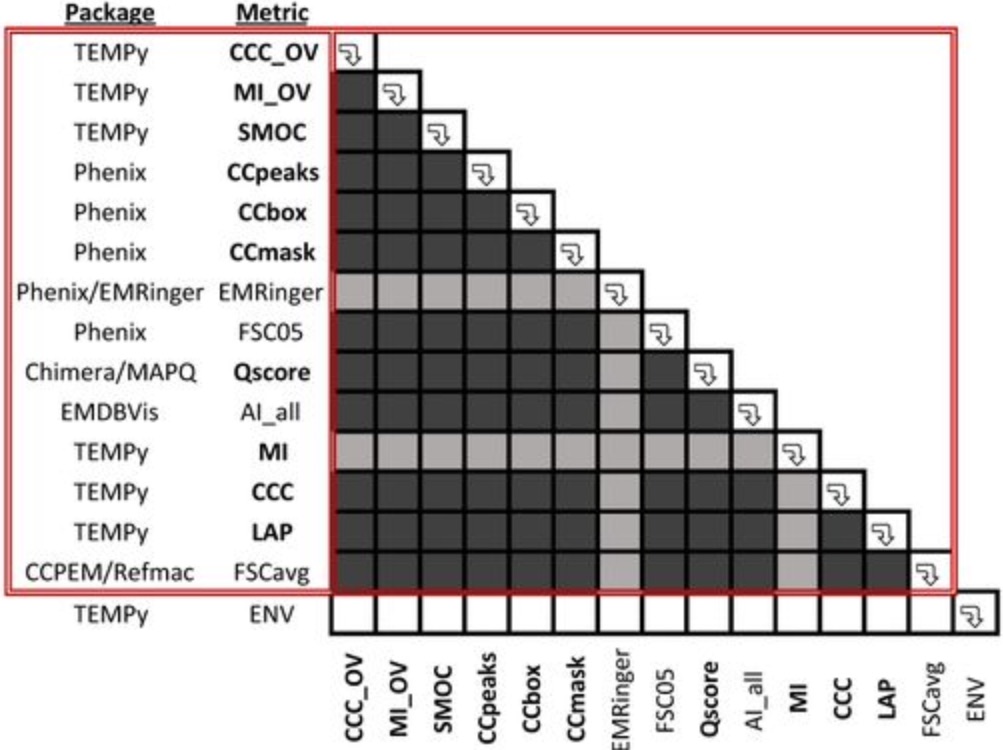
Cryo-EM model validation recommendations based on outcomes of the 2019 EMDataResource challenge.
Lawson CL, Kryshtafovych A, Adams PD, Afonine PV, Baker ML, Barad BA, Bond P, Burnley T, Cao R, Cheng J, Chojnowski G, Cowtan K, Dill KA, DiMaio F, Farrell DP, Fraser JS, Jr. MAH, Hoh SW, Hou J, Hung L, Igaev M, Joseph AP, Kihara D, Kumar D, Mittal S, Monastyrskyy B, Olek M, Palmer CM, Patwardhan A, Perez A, Pfab J, Pintilie GD, Richardson JS, Rosenthal PB, Sarkar D, Schäfer LU, Schmid MF, Schröder GF, Shekhar M, Si D, Singharoy A, Terashi G, Terwilliger TC, Vaiana A, Wang L, Wang Z, Wankowicz SA, Williams CJ, Winn M, Wu T, Yu X, Zhang K, Berman HM, Chiu W. Nature Methods. 2021.
-
Access the paper
- PMID: 33542514
- PMCID: PMC7864804
- Biorxiv Preprint: 147033
- Full Text Additional Links:
- 2019 Model Metrics Challenge
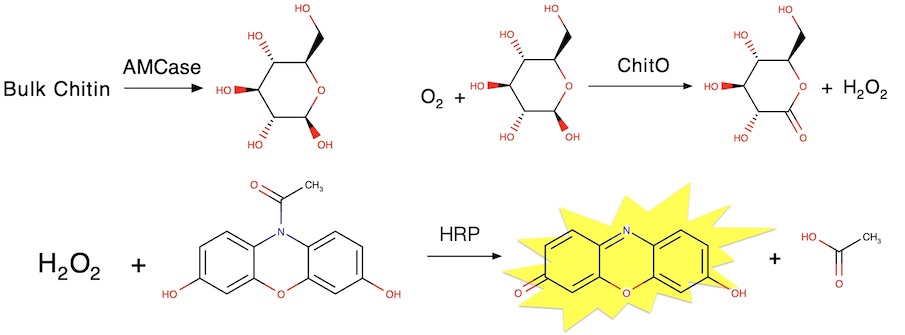
Differences in the chitinolytic activity of mammalian chitinases on soluble and insoluble substrates
Barad BA, Liu L, Diaz RE, Basillo R, Van Dyken SJ, Locksley RM, Fraser JS. Protein Science. 2020.
-
Access the paper
- PMID: 31930591
- PMCID: PMC7096708
- Biorxiv Preprint: 762336
- Full Text Additional Linkss:
- Chitin Analysis Scripts on GitHub
- Relax Scripts for calculating non-linear relaxation fits on GitHub
- Locksley lab @ UC San Francisco
- Van Dyken lab @ Washington University in St. Louis
- Celebratory Tweetstorm
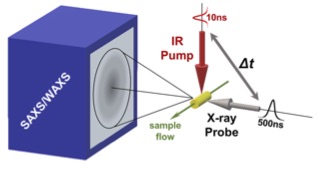
Temperature-Jump Solution X-ray Scattering Reveals Distinct Motions in a Dynamic Enzyme
Thompson MC, Barad BA, Wolff AM, Cho HS, Schotte F, Schwarz DMC, Anfinrud P, Fraser JS. Nature Chemistry. 2019.
-
Access the paper
- PMID: 31527847
- PMCID: In Process
- Biorxiv Preprint: 476432
- Full Text Additional Linkss:
- Solution Scattering Code on Github
- Solution Scattering Data on NIH Figshare
- BioCARS
- Philip Anfinrud
- Paper Submission Celebration Tweetstorm
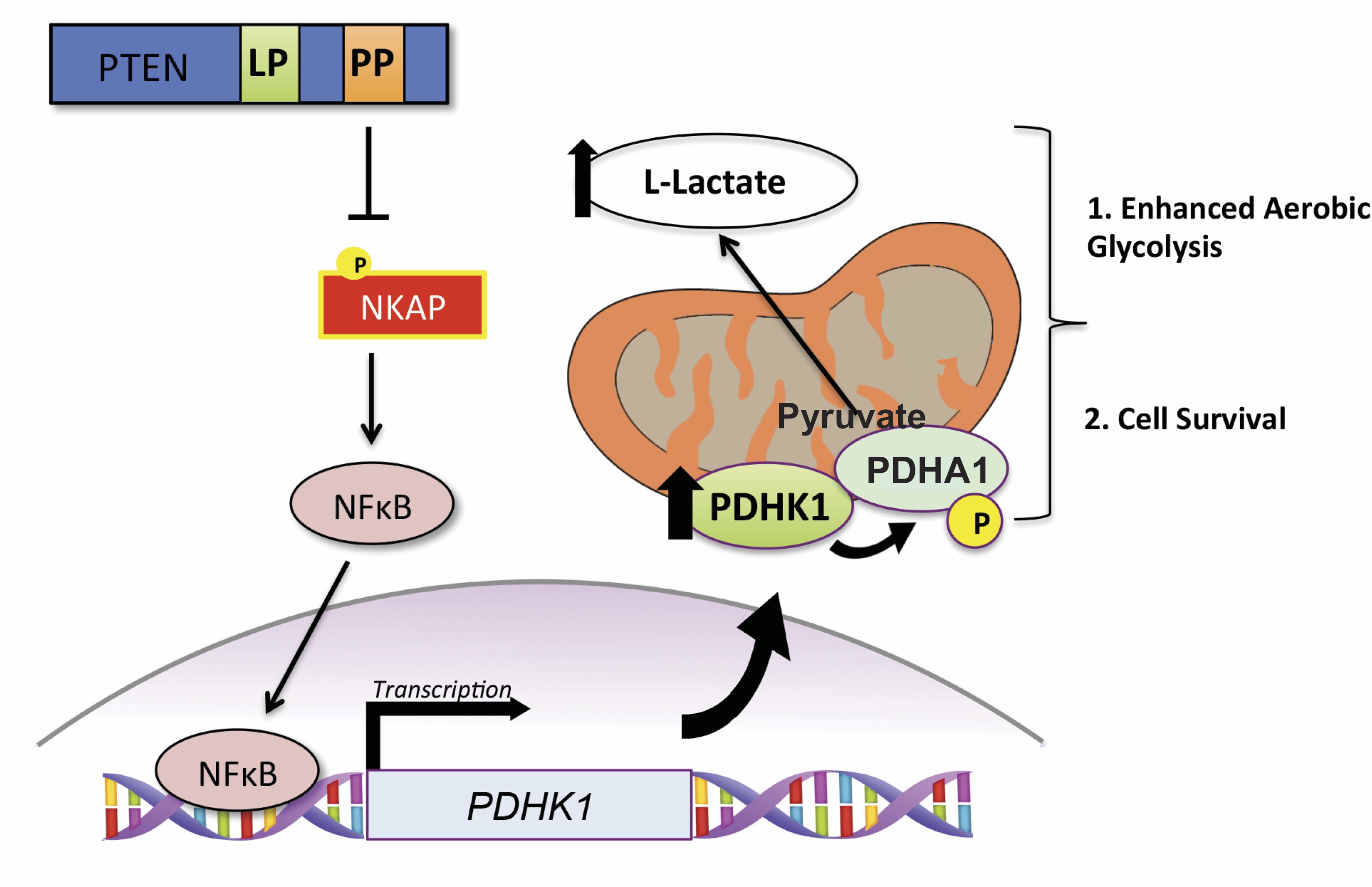
Synthetic essentiality of metabolic regulator PDHK1 in PTEN-deficient cells and cancers
Chatterjee N, Pazarentzos E, Mayekar MK, Gui P, Allegakoen DV, Hrustanovic G, Olivas V, Lin L, Verschueren E, Johnson JR, Hofree M, Yan JJ, Newton BW, Dollen JV, Earnshaw CH, Flanagan J, Chan E, Asthana S, Ideker T, Wu W, Suzuki J, Barad BA, Kirichok Y, Fraser JS, Weiss WA, Krogan NJ, Tulpule A, Sabnis AJ, Bivona TG. Cell Reports. 2019.
-
Access the paper
- PMID: 31461649
- PMCID: PMC6728083
- Biorxiv Preprint: 441295
- Full Text Additional Links:
- Bivona Lab
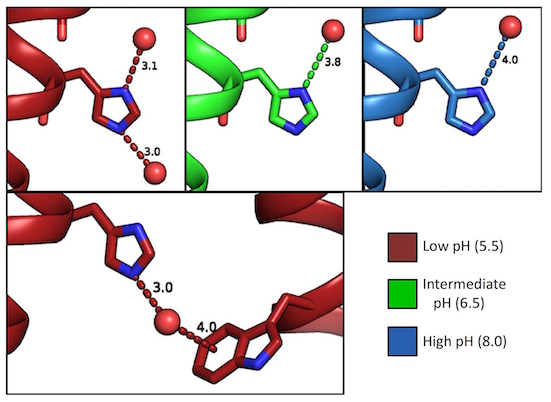
XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction
Thomaston JL, Woldeyes RA, Nakane T, Yamashita A, Tanaka T, Koiwai K, Brewster AS, Barad BA, Chen Y, Lemmin T, Uervirojnangkoorn M, Arima T, Kobayashi J, Masuda T, Suzuki M, Sugahara M, Sauter NK, Tanaka R, Nureki O, Tono K, Joti Y, Nango E, Iwata S, Yumoto F, Fraser JS, DeGrado WF. Proceedings of the National Academy of Sciences. 2017.
-
Access the paper
- PMID: 28835537
- PMCID: PMC5754760
- Full Text
- Deposited Structures: 5JOO, 5UM1, 5TTC Additional Linkss:
- DeGrado Lab
- SACLA XFEL
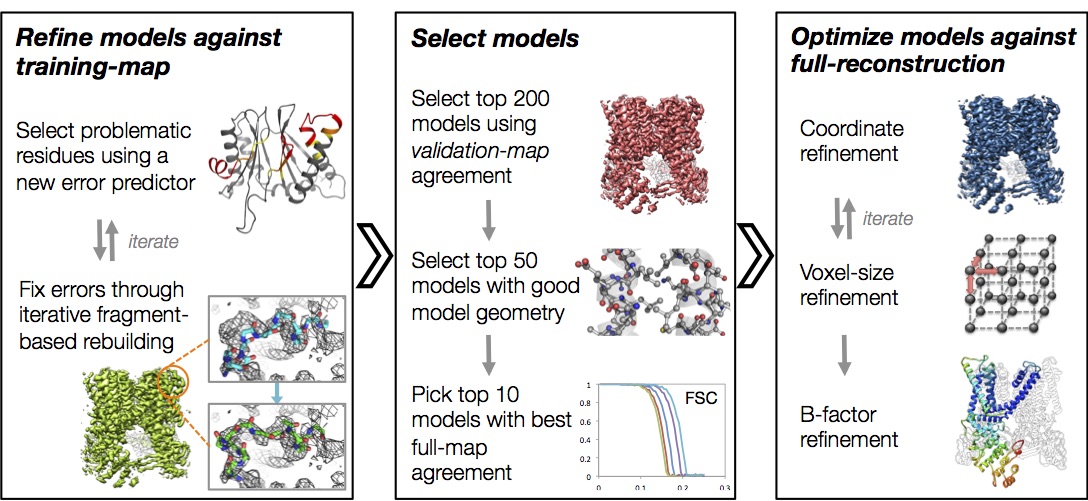
Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta
Wang RYR, Song Y, Barad BA, Cheng Y, Fraser JS, DiMaio F. eLife. 2016.
-
Access the paper
- PMID: 27669148
- PMCID: PMC5115868
- Biorxiv Preprint: 050286
- Full Text Additional Links:
- DiMaio Lab
Determination of Ubiquitin Fitness Landscapes Under Different Chemical Stresses in a Classroom Setting
Mavor D, Barlow KA, Thompson S, Barad BA, Bonny AR, Cario CL, Gaskins G, Liu Z, Deming L, Axen SD, Caceres E, Chen W, Cuesta A, Gate R, Green EM, Hulce KR, Ji W, Kenner LR, Mensa B, Morinishi LS, Moss SM, Mravic M, Muir RK, Niekamp S, Nnadi CI, Palovcak E, Poss EM, Ross TD, Salcedo E, See S, Subramaniam M, Wong AW, Li J, Thorn KS, Conchúir SÓ, Roscoe BP, Chow ED, DeRisi JL, Kortemme T, Bolon DN, Fraser JS. eLife. 2016.

-
Access the paper
- PMID: 27111525
- PMCID: PMC4862753
- Biorxiv Preprint: 025452
- Full Text Additional Linkss:
- Paper Submission Celebration Photo
- PUBS Code on Github
- Course Syllabus
- eLIFEdigest: Yeast in a class of their own
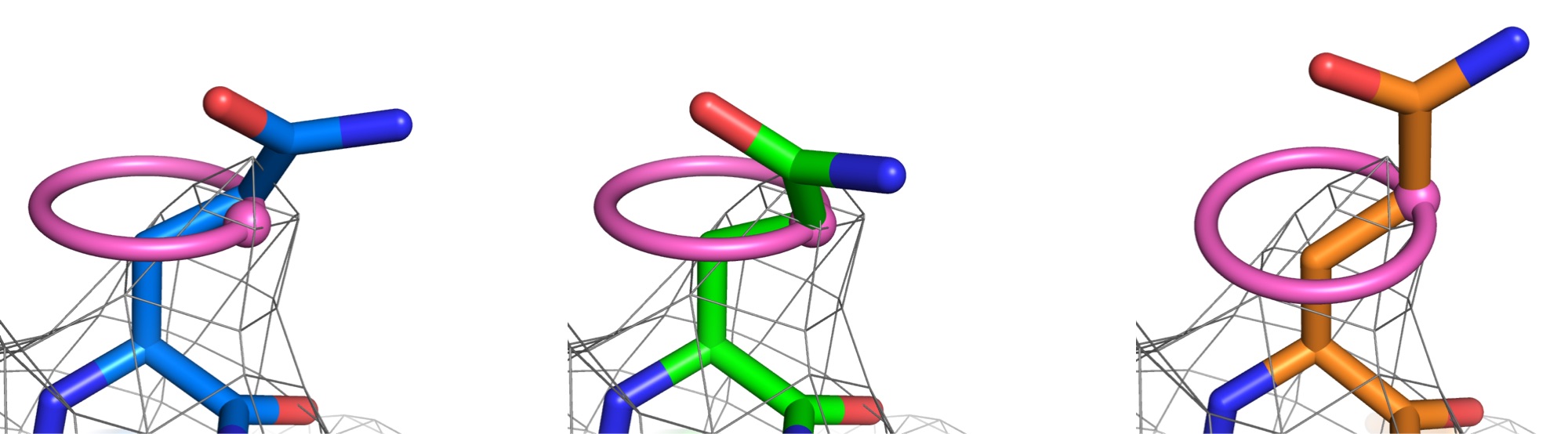
EMRinger: Side-chain-directed model and map validation for 3D Electron Cryomicroscopy
Barad BA, Echols N, Wang RYR, Cheng Y, DiMaio F, Adams PD, Fraser JS. Nature Methods. 2015.
-
Access the paper
- PMID: 26280328
- PMCID: In process
- Biorxiv Preprint: 014738
- Full Text
- Deposited Structures: 3J9I, 3J9J Additional Linkss:
- Paper Submission Celebration Photo
- EMRinger Code on Github
- Blog Post about EMRinger